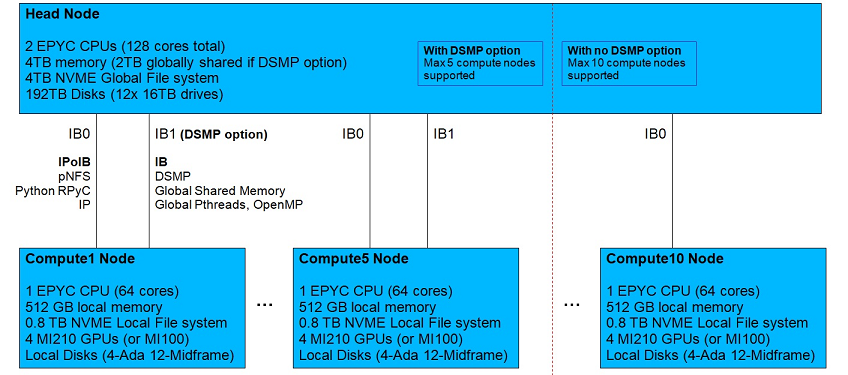
Symmetric Computing creates departmental supercomputers and departmental mainframes that support high performance computing for research and development, engineering simulation, advanced business processes, Machine Learning, Artificial Intelligence, large databases and other computationally and data intensive applications. The Ada computer architecture utilizes a tightly integrated cluster architecture that is directly connected using high speed Infiniband/RDMA. Each system consists of a head node that directly connects to multiple compute/worker nodes, which support GPUs and other accelerators. The head node provides a single system interface that includes a global NVMe file system, a distributed Python runtime environment and an integrated disk array. Ada systems utilize the Linux operating system, and optionally support distributed symmetric multiprocessing (DSMP), an operating system extension that provides applications with global shared memory, pthreads and OpenMP across all nodes. A generalized view of the Ada architecture is illustrated in the following diagram.
Symmetric Multi-Processing (SMP) computers with large shared memory and many processor cores are perfect for Big Data and high-performance computing applications that utilize large in-memory datasets. This includes large in-memory database installations and other critical enterprise software. Symmetric Computing has developed large SMP computers that are a fraction of the cost of currently available mainframe computers using our Distributed Symmetric Multiprocessing (DSMP) technology. The DSMP Linux kernel transforms the Ada cluster into a shared memory, many core supercomputer. A process launched on the head node can take advantage of the memory and cores on worker nodes by using the DSMP supplied Pthreads or OpenMP. To programmers, the system presents as a single large memory Linux box with hundreds or thousands of cores. Large datasets can be read into memory and accessed directly. Often programmers can avoid message passing interface (MPI) and complex file-access programming. This makes it simpler to port applications to a supercomputer.
DSMP adds two key extensions to the Linux kernel. They are distributed shared memory (DSM) and a new kernel based global Pthreads implementation. DSM utilizes a two tiered memory organization, consisting of a local memory partition on each node, and an additional global memory partition on a subset of nodes. The global memory partitions combine to form a single global memory that is addressable by every node. Global memory pages are swapped into local memory partitions on nodes as needed by executing program threads. Modified pages are copied back to global memory. The consistency of global memory is maintained with the aid of a set of memory page locking functions that are available to application programs. A generalized view of this architecture is illustrated in the following diagram.
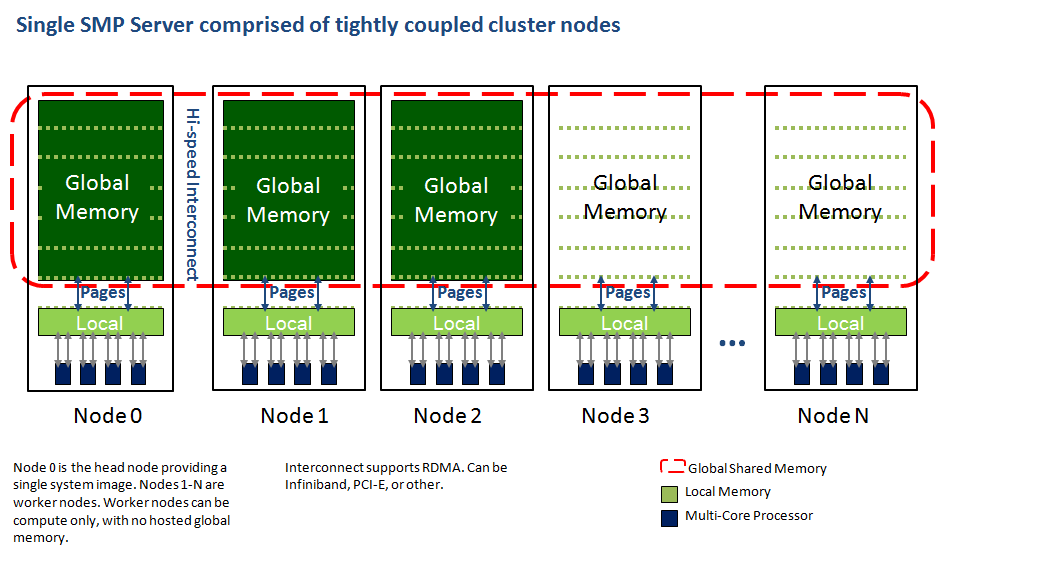
The DSMP Linux kernel extenstions provide large shared memory, many core SMP computing with both economy and performance.
Download the White Paper describing our DSMP Linux Kernel Extension.
The Virtual Drug Discovery Platform (VDDP) is a software pipeline that uses a combination of ligand docking and molecular dynamics to identify small molecule drug candidates. The VDDP is hosted on our Ada hybrid CPU/GPU supercomputer. It includes a database of nearly two billion small molecules with their 3d structures, a database of proteins with their 3d Xray crystal structures from human and other organisms, and a database of over ten thousand experimental and deployed drugs along with their target proteins and binding sites. Using this platform we are able to conduct early stage drug discovery research that significantly reduces the time and cost of identifying lead compounds.
